AsianScientist (Dec. 23, 2014) – Scientists have isolated the gene controlling cotton fiber elgonation, GhHOX3. The finding, published in Nature Communications, could play a role in the future selective breeding of cotton plants, thereby impacting the billion-dollar industry.
Cotton is a major cash crop widely planted in over 80 countries, with an annual output value of US$12 billion, mainly for fiber. Cotton fibers, the most important natural material for textile industry, are extremely long, single-celled seed-born trichomes (hairs).
A research group led by Professor Chen Xiaoya from the Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences set out the identify the genes controlling the development of cotton fibers. In the model plant Arabidopsis thaliana, homeodomain-leucine zipper (HD-ZIP) transcription factors are known to positively regulate the development of trichomes. Previous work by the Chen group found that three HD-ZIP factors are also expressed in cotton plants, namely HOX1, 2 and 3.
In the present study focused on GhHOX3, the researchers found that the gene is associated with quantitative trait loci (QTLs) for fiber length. Silencing of GhHOX3 dramatically reduces fiber length, whereas its overexpression leads to longer fibers.
The team went on to show that GhHOX3 regulates target genes—including the cell wall loosening protein genes GhRDL1 and GhEXPA1—by binding to the L1-box in the promoter. GhHOX3 also interacts with GhHD1, another homeodomain protein, resulting in enhanced transcriptional activity, as well as with cotton DELLA protein GhSLR1, a major repressor of the growth hormone gibberellin (GA). GA enhances GhHOX3 activity and fiber cell growth through degrading GhSLR1.
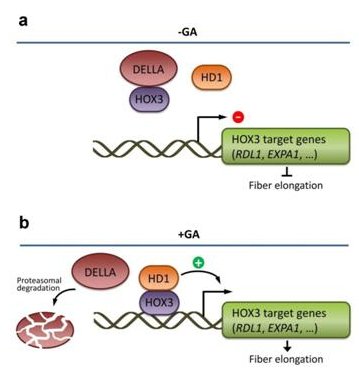
This study has identified a key regulator of cotton fiber elongation for the first time. Moreover, the direct interaction between DELLA and a specific homeodomain protein sheds new light on the biochemical mechanism of GA—a widely acknowledged green evolution hormone—in controlling plant cell elongation.
The article can be found at: Shan et al. (2014) Control of Cotton Fibre Elongation by a Homeodomain Transcription Factor GhHOX3.
———
Source: Shanghai Institutes for Biological Sciences.
Disclaimer: This article does not necessarily reflect the views of AsianScientist or its staff.