AsianScientist (Sep. 4, 2015) – Scientists have developed a mathematical model that predicts how biological circuits generate rhythms and control their robustness. This work, published in Science, is a result of a collaboration between Rice University, University of Houston and Korea Advanced Institute for Science and Technology (KAIST).
Our bodies have a variety of biological clocks that follow rhythms or oscillations with periods ranging from seconds to days. For example, our hearts beat every second and cells divide periodically. The circadian clock located in the hypothalamus generates twenty-four hour rhythms, timing our sleep and hormone release. How do these biological clocks or circuits generate and sustain the stable rhythms that are essential to life?
A team of researchers led by Dr. Matthew Bennett of Rice University set out to answer this question using synthetic biology. The top-down research approach, which focuses on identifying the components of biological circuits, limits our understanding of the mechanisms in which the circuits generate rhythms. Synthetic biology, a rapidly growing field at the interface of biosciences and engineering, however, uses a bottom-up approach.
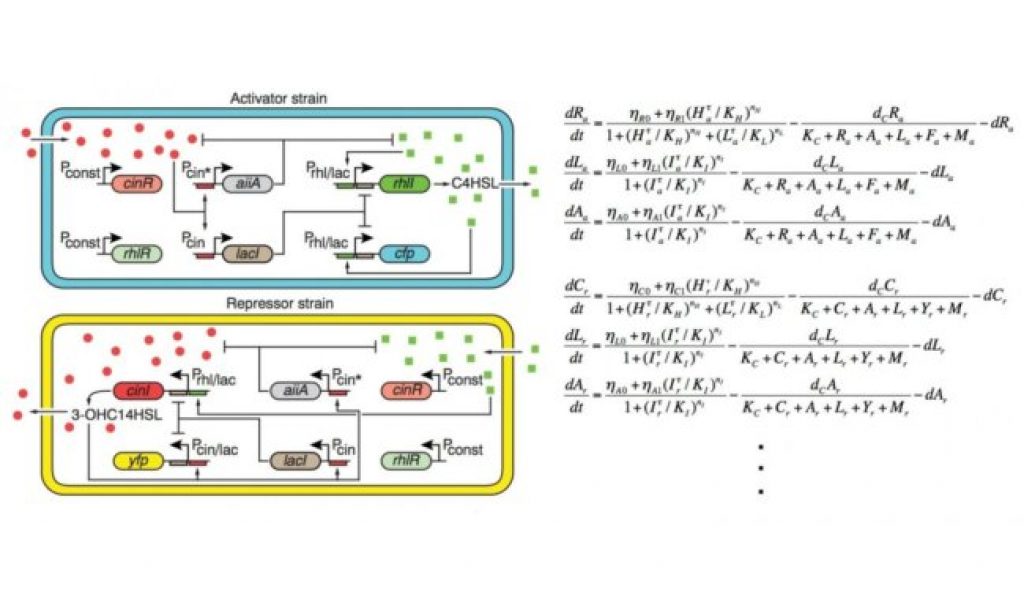
They constructed a novel biological circuit that spans two genetically engineered strains of bacteria, one serving as an ‘activator’ and the other as a ‘repressor’ to regulate gene expression across multiple cell types. They found that the circuit generates surprisingly robust rhythms under various conditions.
As suggested by the mathematical analysis of co-first author Kim Jae Kyong from KAIST and confirmed experimentally, the presence of negative feedback loops in addition to a core transcriptional negative feedback loop can explain the robustness of rhythms in this system. This result provides important clues about the fundamental mechanism of robust rhythm generation in biological systems.
Furthermore, rather than constructing the entire circuit inside a single bacterial strain, the circuit was split among two strains of Escherichia coli bacterium. When the strains were grown together, the bacteria exchanged information, completing the circuit. Thus, this research also shows how, by regulating individual cells within the system, complex biological systems can be controlled, which in turn influences each other (e.g., the gut microbiome in humans).
The article can be found at: Chen et al. (2015) Emergent Genetic Oscillations in a Synthetic Microbial Consortium.
———
Source: Korea Advanced Institute of Science and Technology.
Disclaimer: This article does not necessarily reflect the views of AsianScientist or its staff.